Elias Dohmen
Tools for Data Analysis and Biological Discovery
This page presents a selected set of bioinformatic tools that I have developed or contributed to. These tools span a range of biological topics and are designed to support researchers in analysing their own data and addressing their research questions. The focus is on accessibility and broader applicability across the scientific community.
Please note that this is only a representative selection. For a more comprehensive overview of my work and related tools, you are welcome to explore the following platforms:
- GitHub - personal GitHub with my own projects and open-source software
- ZIV GitLab - institutional GitLab with personal projects and collaborative software I contributed to
- DomainWorld - software collection of tools for protein domain analysis and annotation
I am committed to supporting open, reproducible, and transparent research. The tools listed here reflect that commitment and are intended to empower researchers with reliable and reusable resources.
-

DOGMA A tool for fast and easy quality assessment of transcriptome and proteome data based on conserved protein domains.
Implementation: Python
User Interface: CLI, Webserver
Linked Publications: 📄 📄
Availability: 🏠🌐
modern version with additional features in development...stay tuned! -
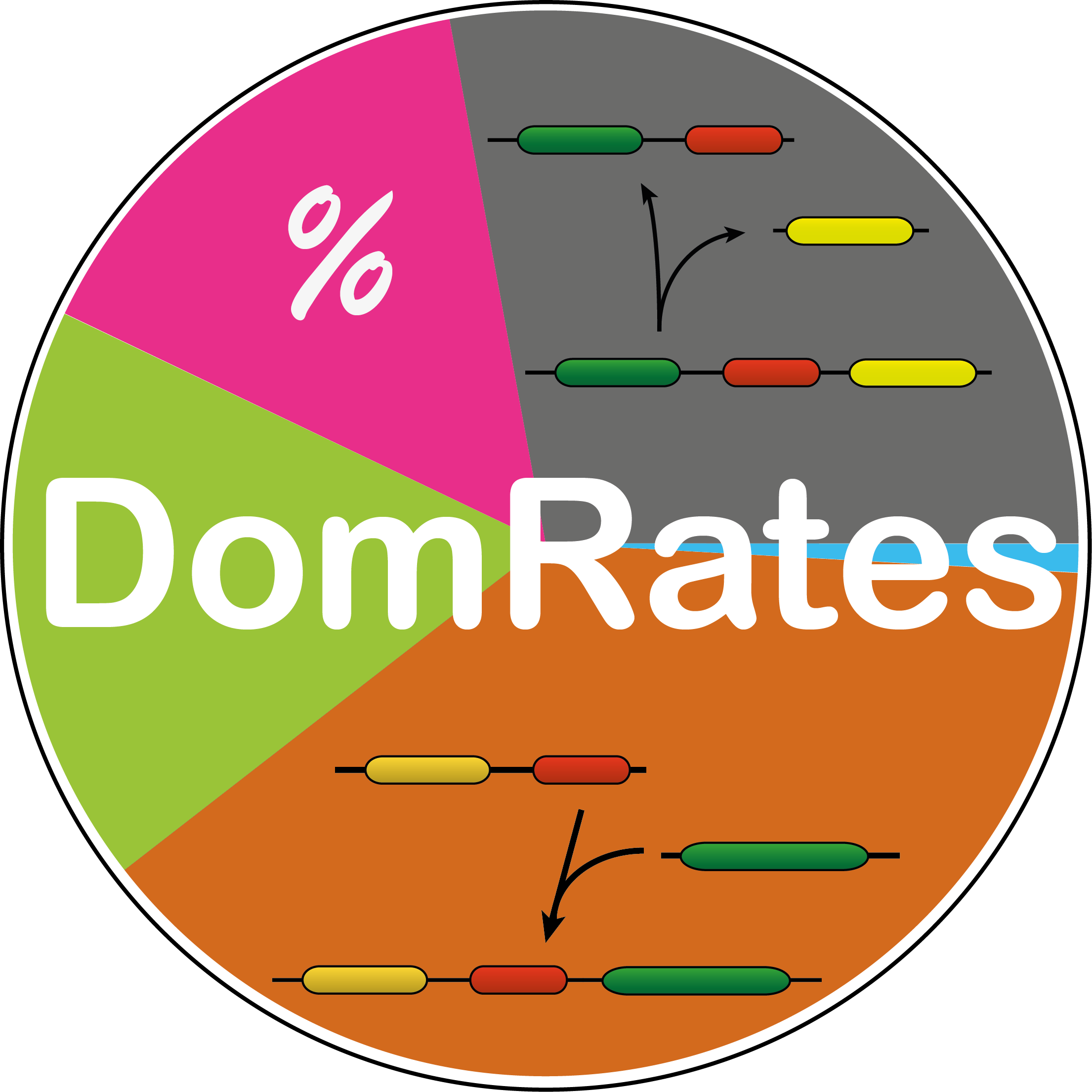
DomRates A tool for infering domain rearrangement events and their rates across phylogenetic trees.
Implementation: C++
User Interface: CLI
Linked Publications: 📄 📄
Availability: 🏠 -

DeNoFo A file format and toolkit for standardized, comparable de novo gene annotation.
Implementation: Python
User Interface: GUI, CLI
Linked Publications: 📄 📄
Availability: 🏠🐍
-

DESWOMAN A tool for automatic detection of de novo gene precursors and analysis of their mutations based on transcriptome data.
Contribution: Python packaging and dockerizing, consulting
Implementation: Python
User Interface: GUI, CLI
Linked Publications: 📄
Availability:🐳
-
GEvol pipeline A Snakemake-based bioinformatic workflow designed to streamline the evolutionary analyses in comparative proteomics.
Implementation: Snakemake
User Interface: CLI
Linked Publications: coming soon...
Availability: in development -

DomTree A Deep Learning-based tool to reconstruct phylogenetic trees based on whole proteome information through protein domains.
Implementation: Python, C++
User Interface: CLI
Linked Publications: Chapter 5 in: 📄 Availability: in development
A new improved version, addressing the described problems and shortcomings in the linked publication is currently in development.
